Our Mission
To develop an information capability at the forefront of personalized cancer treatments. We achieve this through the assembly of experimental scaffolds that help us understand tumor complexity and predict clinical outcomes.

We conduct large-scale projects that capture the diversity of our patients and provide a rich substrate for computational and mathematical models of cancer’s propensity to resist our treatments.
Deep Learning Our Patients’ Clinomic Networks
Clinomics is the integration of clinical information (e.g. demographics, treatments, outcomes) with omic outputs for individual patients. The latter include radiomics (embedded quantitative data derived from imaging modalities like computed tomography or magnetic resonance), genomics (genetic information derived from the patient’s tumor or germline), transcriptomics (gene expression), and others. We recently showed that an image- and clinical-based deep learning framework can predict treatment failure after radiotherapy to the lung. We seek to study model transportability (e.g. across hospital systems, patient demographics and ethnicity, image scanners), interpretability (what is the basis that the machine uses to make these predictions?), and model stability overtime (adaptive models in a dynamic population). We seek to design and implement devices that can augment the physician’s ability to estimate the probability of treatment failures and modulate failure by individualized treatment recommendations.
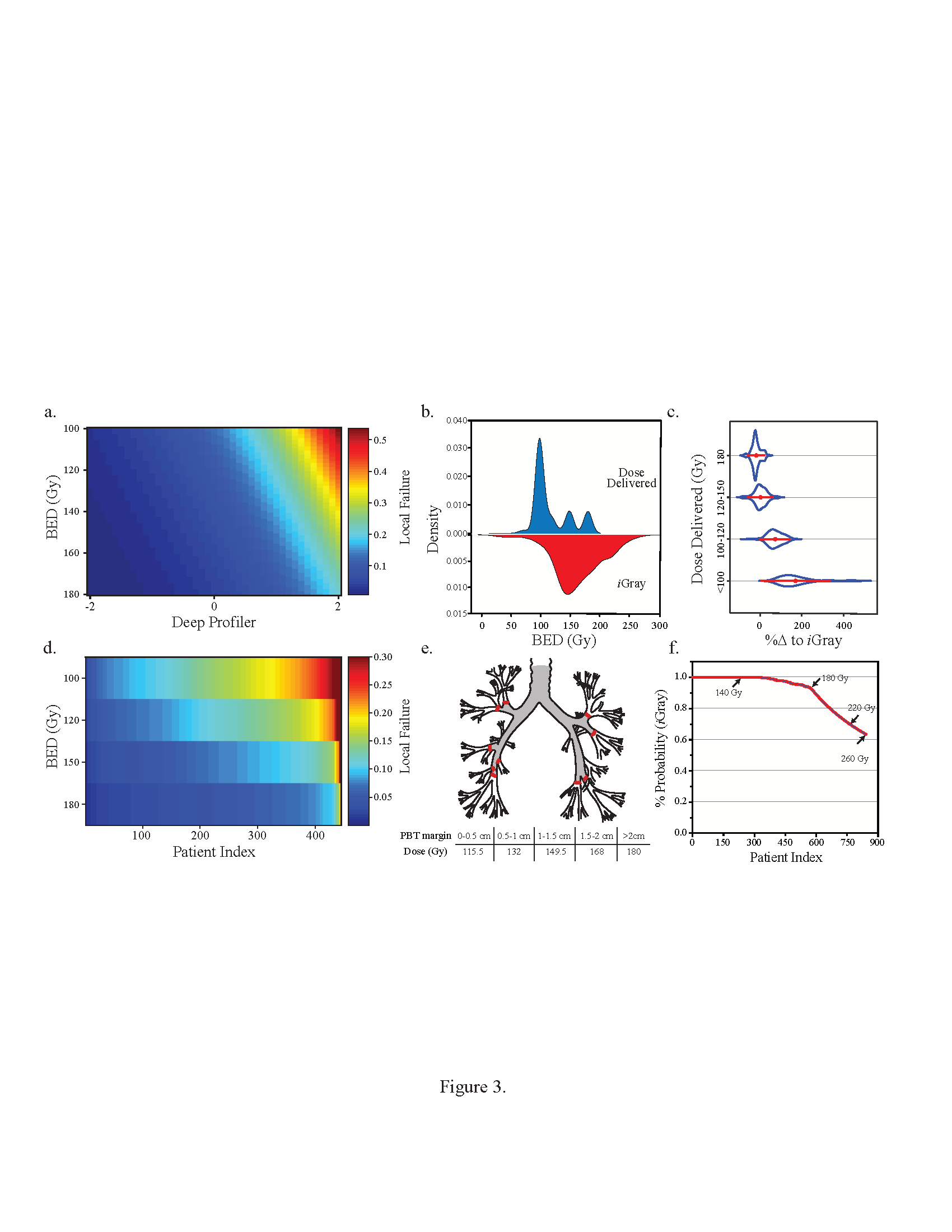
Stochastic Transitions and Phenotypic Equilibria
We recently identified distinct and commutative cancer states that confer complimentary functional attributes in individual small cell lung carcinoma, establishing a dynamic ecosystem that promotes tumor survival under environmental and/or therapeutic stress. We combined high-resolution measurement of cell state dynamics at the single cell level, invocation of stochastic transition theory to estimate transition probabilities using a discrete-time Markov model, and correlative studies using human-derived xenografts and clinical outcomes in order to better characterize these states, their dynamics, and their putative roles in modulating clinical outcomes. We showed that individual lung tumors display distinctive equilibria in the proportion of cells in these well-delimited, transcription factor defined cellular states (A, Ascl1; N, NeuroD1; Y, Yap1). We also show that phenotypic switching and transcriptional reprogramming represent critical mechanisms for tumor persistence during chemotherapy, ultimately leading to therapeutic resistance and aggressive tumor recurrence. We are currently elucidating the molecular mechanisms of these ‘switches” and we seek to identify new therapeutic targets to combat plasticity in this highly lethal lung cancer subtype.
PDXray
The 10,000 Avatar Project was inaugurated by our group in the last quarter. This will be the largest patient-derived xenograft (PDX) mouse experiment conducted to date by any group. ~10,000 mice engrafted with ~500 genetically annotated PDXs will be irradiated using a singular experimental platform. This work will correlate genetic and other omic (e.g. transcriptomic, metabolomic, et cetera) alterations with the likelihood of response to radiotherapy and matched recurrent tumors.

Atlas
The Pan-cancer Radiogenomic Atlas is a gene variant profiling project that seeks to interrogate >1000 common and rare genetic variants for response to ionizing radiation in immortalized human cells (non-cancer cells). It uses novel CRISPR methodology to examine the impact of individual cancer variants on the likelihood of survival after therapy. The initial phase was completed in December, 2019. Current work is building on the unary profiling methodology to study the interaction between varied gene variants, thus building toward greater complexity. The latter will allow us to model non-linear, multi-variable systems in a manner that is amenable to experimental manipulation and validation. Whereas the PDXray project is a top-down profiling project (using human tissue in murine systems to deconvolve biomarkers of response to radiotherapy), Atlas seeks to reconstruct tumors and their complexity, a single mutation at a time.

XTD2
The X-ray Target Discovery and Development (XTD2) project profiled 533 cancer cell lines comprising 26 cancer types to ionizing radiation. Published in 2016, this project represented the largest profiling effort of cancer cell line survival after irradiation ever conducted. Our results demonstrated significant biological diversity in the survival of cancer cells to ionizing radiation and established multiple lines of evidence that new genetic features regulating cellular response after DNA damage can be identified.

